library(bigrquery) # R Interface to Google BigQuery API
library(dplyr) # Grammar for data manipulation
library(DBI) # Interface definition to connect to databases RStudio Setup
Overview
dplyr as a database interface
This information was found here. I refer to that link to connect to DBs.
More information can be found at dplyr regarding connecting to db and lazy evaluation.
This page is just an overview of why I chose RStudio and doesn’t show the intricate details of connections. The dplyr package simplifies data transformation. It provides a consistent set of functions, called verbs, that can be used in succession and interchangeably to gain understanding of the data iteratively.
dplyr is able to interact with databases directly by translating the dplyr verbs into SQL queries. This convenient feature allows you to ‘speak’ directly with the database from R. Other advantages of this approach are:
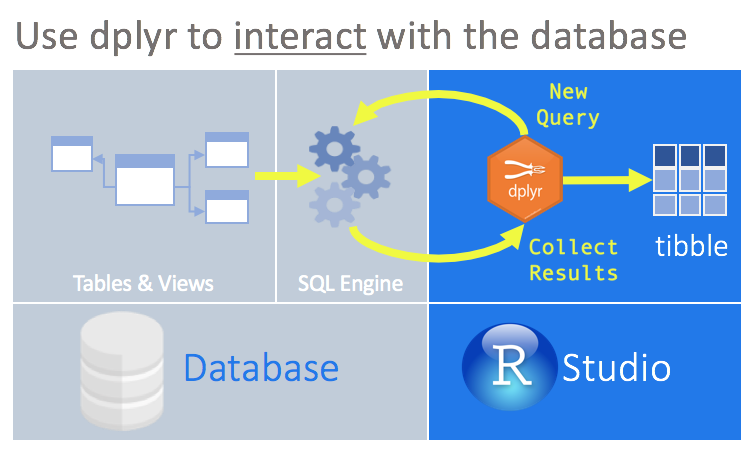
- Run data exploration routines over all of the data, instead of importing part of the data into R.
- Use the SQL Engine to run the data transformations. In effect, computation is being pushed to the database.
- Collect into R only a targeted dataset.
- All of your code is in R. Because
dplyris used to communicate with the database, there is no need to alternate between languages or tools to perform the data exploration.
Connect to DB
At the center of this approach is the DBI package. This package acts as ‘middle-ware’ between packages to allow connectivity with the database from the user or other packages. It provides a consistent set of functions regardless of the database type being accessed. The dplyr package depends on the DBI package for communication with databases.
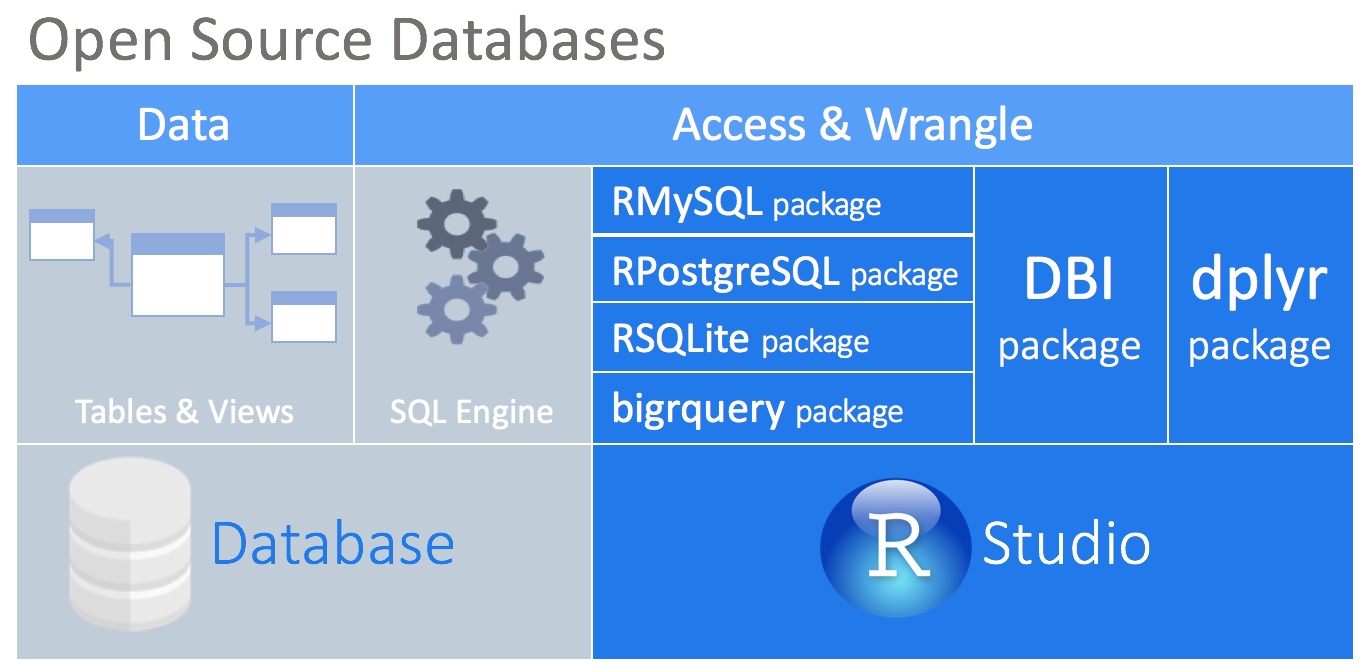
There are packages that enables a direct connection between the an open-source database and R. Currently, such packages exist for the following databases: MySQL, SQLite, PostgreSQL, and BigQuery.
Most commercial databases, like Oracle and Microsoft SQL Server, offer ODBC drivers that allow you to connect your tool to the database. Even though there are R packages that allow you to use ODBC drivers, the connection will most likely not be compatible with DBI. The new odbc package solves that problem by providing a DBI backend to any ODBC driver connection.
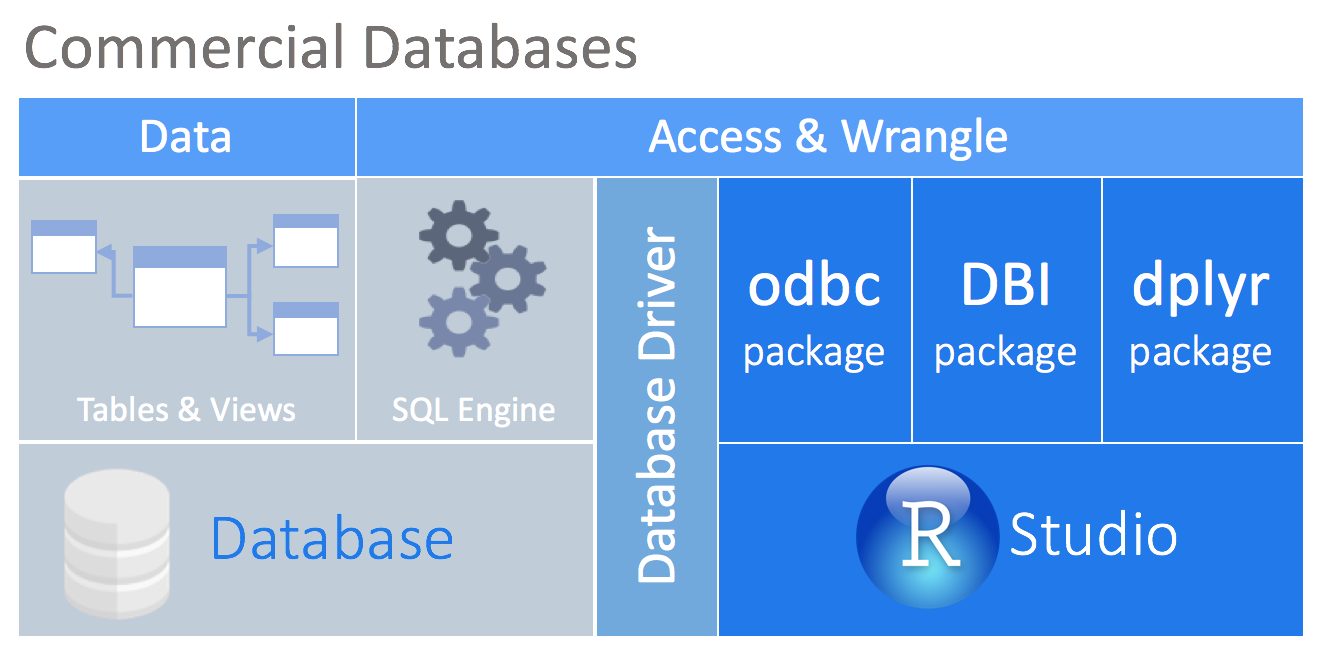
SQL Translations for dplyr
A complementary package called dbplyr contains the translations of the vendor-specific SQL for dplyr to use. A list of known supported databases are available in our Databases page.
Here is a breakdown of the DBs
Databases
| Name | Posit Pro Driver | dplyr support | Connect via R package |
|---|---|---|---|
| Amazon Redshift | ✅ | ✅ | |
| Apache Hive | ✅ | ✅ | |
| Apache Impala | ✅ | ✅ | |
| Athena | ✅ | ||
| Cassandra | ✅ | ||
| Databricks | ✅ | ✅ | |
| Google BigQuery | ✅ | ✅ | bigrquery |
| Microsoft SQL Server | ✅ | ✅ | |
| MonetDB | ✅ | MonetDBLite | |
| MongoDB | ✅ | ||
| MySQL | ✅ | ✅ | RMariaDB |
| Netezza | ✅ | ||
| Oracle | ✅ | ✅ | |
| PostgreSQL | ✅ | ✅ | RPostgres |
| SQLite | ✅ | RSQLite | |
| Salesforce | ✅ | ||
| Snowflake | ✅ | ✅ | |
| Teradata | ✅ | ✅ |
Connect to BigQuery
As we know that R loads data into memory which could be taxing with large datasets. In this section I’ll connect to Google BigQuery and access some of their large public datasets.
I already have an account setup in BigQuery as I’ve done lots of work on it, and I also have some sample projects that I saved on Google Cloud that we’ll access in this section to demonstrate SQL from within RStudio and how to integrate R and SQL in the same analysis.
What follows is a short example on how to connect to BigQuery, access a project, data, query the data with R and SQL all using RStudio desktop which is free.
Packages
- “bigrquery”, “dplyr” and “DBI” packages provide an abstraction over the underlying BigQuery REST API to interact with the data.
- bigrquery and DBI provide a low level wrapper over the BigQuery API’s whereas dplyr provides a higher level of abstraction and lets BigQuery tables be treated as data frames.
Setup the Connection
library(bigrquery) # R Interface to Google BigQuery API
library(dplyr) # Grammar for data manipulation
library(DBI)
projectid <- 'prime-depot-415622'
datasetid <- 'BikeShare'
con <- dbConnect((bigquery()),
project = projectid,
dataset = datasetid,
use_legacy_sql = FALSE)List Datasets in Project
SELECT *
EXCEPT(schema_owner, location, DDL, default_collation_name)
FROM
INFORMATION_SCHEMA.SCHEMATA | catalog_name | schema_name | creation_time | last_modified_time | sync_status |
|---|---|---|---|---|
| prime-depot-415622 | cus_data | 2024-03-03 21:24:36 | 2024-03-03 21:24:36 | NULL |
| prime-depot-415622 | bellabeat | 2024-03-20 20:06:32 | 2024-03-20 20:06:32 | NULL |
| prime-depot-415622 | employee_data | 2024-03-06 23:59:41 | 2024-03-06 23:59:41 | NULL |
| prime-depot-415622 | demos | 2024-03-05 17:43:26 | 2024-03-05 17:43:26 | NULL |
| prime-depot-415622 | cars | 2024-03-03 18:17:34 | 2024-03-03 18:17:34 | NULL |
| prime-depot-415622 | babynames | 2024-02-28 17:06:55 | 2024-02-28 17:06:55 | NULL |
| prime-depot-415622 | BikeShare | 2024-04-01 20:47:30 | 2024-04-01 20:47:30 | NULL |
| prime-depot-415622 | city_data | 2024-02-28 16:34:15 | 2024-02-28 16:34:15 | NULL |
| prime-depot-415622 | customer_data | 2024-03-03 16:27:32 | 2024-03-03 16:27:32 | NULL |
| prime-depot-415622 | nfl | 2024-03-04 17:32:41 | 2024-03-04 17:32:41 | NULL |
List Tables w/R
To view the tables in the DB use dbListTables()
dbListTables(con)[1] "trips19v1" "trips20" List Tables w/SQL
SELECT table_catalog, table_schema, table_name, table_type, is_insertable_into
FROM BikeShare.INFORMATION_SCHEMA.TABLES| table_catalog | table_schema | table_name | table_type | is_insertable_into |
|---|---|---|---|---|
| prime-depot-415622 | BikeShare | trips19v1 | BASE TABLE | YES |
| prime-depot-415622 | BikeShare | trips20 | BASE TABLE | YES |
List Column Schema
SELECT table_name, column_name, ordinal_position, is_nullable, data_type
FROM BikeShare.INFORMATION_SCHEMA.COLUMNS| table_name | column_name | ordinal_position | is_nullable | data_type |
|---|---|---|---|---|
| trips19v1 | ride_id | 1 | YES | INT64 |
| trips19v1 | started_at | 2 | YES | TIMESTAMP |
| trips19v1 | ended_at | 3 | YES | TIMESTAMP |
| trips19v1 | ride_length | 4 | YES | STRING |
| trips19v1 | rideable_type | 5 | YES | INT64 |
| trips19v1 | tripduration | 6 | YES | FLOAT64 |
| trips19v1 | start_station_id | 7 | YES | INT64 |
| trips19v1 | start_station_name | 8 | YES | STRING |
| trips19v1 | end_station_id | 9 | YES | INT64 |
| trips19v1 | end_station_name | 10 | YES | STRING |
tbl
Access Tables
As mentioned earlier dplyr allows access to BigQuery tables and allow us to treat them as dataframes
- use dplyr::tbl() to access the tables
- arguments: connection and TableName
Access and assign tables
trips_19 <- tbl(con, "trips19v1")
trips_20 <- tbl(con, "trips20") |> show_query()<SQL>
SELECT *
FROM `trips20`- As you see the SQL query is all printed out for you
class(trips_19)[1] "tbl_BigQueryConnection" "tbl_dbi" "tbl_sql"
[4] "tbl_lazy" "tbl" Note: trips_19 & trips_20 are of class tbl_sql and tbl_lazy. So we have a direct reference to the table in BigQuery but we have not brought the data into memory
Query with R
Let’s use dplyr verbs to access the data in BigQuery, with a simple case of counting the number of all users in the groups.
library(gt)
trips_20 |> count(member_casual) |> gt() |>
opt_table_outline()|>
tab_options(column_labels.background.color = '#1a65b9')| member_casual | n |
|---|---|
| casual | 48480 |
| member | 378407 |
trips_19 |> count(member_casual) |>
gt() |>
opt_table_outline() |>
tab_options(column_labels.background.color = '#1a65b9')| member_casual | n |
|---|---|
| Subscriber | 341906 |
| Customer | 23163 |
Show SQL Query
We can see how the R script translate to an SQL query by adding one show_query() into the code
trips_19 |> count(member_casual) |> show_query()
OUTPUT
<SQL>
SELECT `member_casual`, count(*) AS `n`
FROM `trips19v1`
GROUP BY `member_casual`Query with SQL
We’re going to take the SQL query generated above and use it to directly query the database.
All we do is replace {r} with {sql} at the top of the code chunk and
provide the connection = con (defined earlier in the page)
output.var = “mydataframe” will assign the output of the query into “mydataframe” so it can be used in subsequent R code chunks
{sql, connection = con, output.var = "mydataframe"}In this case we didn’t use the third argument “output.var”
{sql, connection = con }
SELECT `member_casual`, count(*) AS `n`
FROM `trips19v1`
GROUP BY `member_casual`| member_casual | n |
|---|---|
| Subscriber | 341906 |
| Customer | 23163 |